The Von Willebrand factor-ADAMTS-13 axis: a two-faced Janus in bleeding and thrombosis
Accepted: 1 April 2022
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Authors
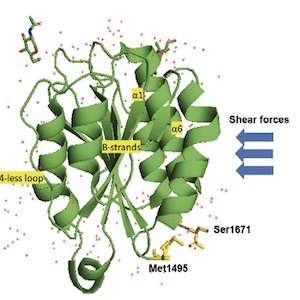
Von Willebrand factor (VWF), a blood multimeric protein with a very high molecular weight, plays a crucial role in the primary hemostasis, the physiological process characterized by the adhesion of blood platelets to the injured vessel wall. Hydrodynamic forces are responsible for the VWF multimers conformational transitions from a globular to a stretched linear conformation. These characteristics render this protein a valuable object to be investigated by mechanochemistry, the biophysical chemistry branch that studies the effects of shear forces on protein conformation. This review will focus on the structural elements of the VWF molecule involved in the biochemical response to shear forces. The stretched VWF conformation favors the interaction with the platelet GpIb and at the same time with ADAMTS-13, the zinc-protease that cleaves VWF in the A2 domain, limiting its prothrombotic capacity. It is important to consider the level or the function of VWF or ADAMTS-13 always in relation each other, keeping in mind that in many thrombotic forms of microangiopathies the reduction of the ratio between the ADAMTS-13 activity and the VWF level (lower than 0.5) can be a valuable parameter to predict a real thrombotic risk. Hence, a significant increase in VWF level alone, even without any reduction of ADAMTS-13 concentration, would still be responsible for a significant reduction of the ADAMTS-13/VWF ratio, which ultimately could reflect or predict a prothrombotic risk. Future studies will have to validate the concept whether ADAMTS-13/VWF ratio could a valuable and reliable biomarker to predict or confirm the presence of thrombotic risk in several morbid conditions.
How to Cite

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
PAGEPress has chosen to apply the Creative Commons Attribution NonCommercial 4.0 International License (CC BY-NC 4.0) to all manuscripts to be published.
Most read articles by the same author(s)
- Giovanni Di Minno, Gaia Spadarella, Ilenia Lorenza Calcaterra, Giancarlo Castaman, Paolo Simioni, Raimondo De Cristofaro, Cristina Santoro, Flora Peyvandi, Matteo Di Minno, The evolving landscape of gene therapy for congenital severe hemophilia: a 2024 state of the art , Bleeding, Thrombosis and Vascular Biology: Vol. 3 No. 2 (2024)
Similar Articles
- Giovanni de Gaetano, Welcome to AICE and sincere wishes for peace for the new year 2024 , Bleeding, Thrombosis and Vascular Biology: Vol. 2 No. 4 (2023)
- Nadim Tawil, Lata Adnani, Janusz Rak, Coagulome and tumor microenvironment: impact of oncogenes, cellular heterogeneity and extracellular vesicles , Bleeding, Thrombosis and Vascular Biology: Vol. 3 No. s1 (2024): 12th ICTHIC
- Pier Mannuccio Mannucci, An ecological alliance against air pollution and cardiovascular disease , Bleeding, Thrombosis and Vascular Biology: Vol. 1 No. 1 (2022)
You may also start an advanced similarity search for this article.

 https://doi.org/10.4081/btvb.2022.11
https://doi.org/10.4081/btvb.2022.11